Last modified on 03 January 2007.
As multiple sequences have become available for different strains of individual organisms, in part due to the increase in genomic sequencing, we can now explore the extent of variation within selected genes from a single species (microheterogeneity).
Sample and Legend:
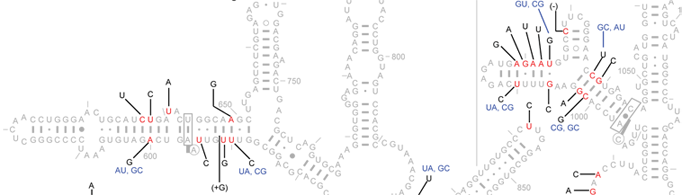
Here, we provide a microheterogeneity analysis of the Escherichia coli 16S rRNAs as a sample. Among the 57 sequences studied here, 90 of the 1542 positions (5.8%) show variations. These 90 positions are shown in red, while all non-variable positions are shown in grey. Black letters reveal the known microheterogeneity at each given position.
When two positions with microheterogeneity are base-paired, the observed base-pair combinations are shown in blue, adjacent to the microheterogeneity descriptions of the 5' nucleotide (space permitting).
Positions marked with "-" and "+" indicate deletions and insertions relative to the reference sequence, respectively.
Microheterogeneity Diagrams:
| RNA Molecule | Organism | # of Sequences | Microheterogeneity Diagram(s) |
|---|---|---|---|
| 16S rRNA | Escherichia coli | 57 | PS | PDF |
