Last modified on 06 June 2014.
| Item | Description | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Attribute | Fields of defined information (abbreviation):
|
|||||||||||||||||||||||||||||||||||
| Organism |
Organisms are most commonly named here with the binomial system of nomenclature, "Genus species." Some organism
names contain subspecies, strain, or operon information.
Examples:
|
|||||||||||||||||||||||||||||||||||
| Phylogenetic Classification, m |
The organism names available here are classified into the three primary phylogenetic domains [Woese C.R., Kandler O., and Wheelis M.L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990 87:4576-4579].
This phylogenetic classification scheme is maintained by the National Center for Biotechnology Information (NCBI), and can be found at their Taxonomy Browser. |
|||||||||||||||||||||||||||||||||||
| Common Name |
The common name database is maintained by the National Center for Biotechnology Information (NCBI), and found at their Taxonomy Browser. Upper case names are specific common names for that "genus species" while lower case are names for a broader class including genus, family or order.
Selected common name groupings have been predefined and made available via the "Animals," "Fungi&Plants," and "Protists" buttons. |
|||||||||||||||||||||||||||||||||||
| L: Cell Location |
cellular location of the RNA
|
|||||||||||||||||||||||||||||||||||
| RT: RNA Type |
Currently, this database actively maintains two RNA types: Introns (primarily group I introns) and Ribosomal RNA (primarily 16S and 23S rRNA). Transfer RNA (tRNA) data will be added in the future.
|
|||||||||||||||||||||||||||||||||||
| RC: RNA Class |
rRNA Classes:
|
|||||||||||||||||||||||||||||||||||
tRNA Classes:
|
||||||||||||||||||||||||||||||||||||
|
The introns available here are divided into groups I and II. The group I introns are further divided into several subgroups, as initially defined by Michel and Westhof [Michel F. and Westhof E. (1990). Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. Journal of Molecular Biology 216:585-610.]
Intron Classes:
|
||||||||||||||||||||||||||||||||||||
| Ex: Exon (the gene in which the intron is located) |
Genes and their abbreviations:
|
|||||||||||||||||||||||||||||||||||
| IN: Intron Number | This number is assigned to an intron to easily distinguish between multiple introns that occur within the same gene. If the gene contains only one intron, the assigned number is 1 (one). | |||||||||||||||||||||||||||||||||||
| IP: Intron Position | This number indicates the intron's position in a 16S or a 23S rRNA molecule (the value refers to the nucleotide number in the corresponding Escherichia coli molecule). This number is helpful for comparing the various occurances of introns between sequences. | |||||||||||||||||||||||||||||||||||
| Size: Sequence Length | Length of the sequence (in nucleotides). [NOTE: only sequences that are more than 90% complete and publicly available are currently included in this RDBMS.] | |||||||||||||||||||||||||||||||||||
| Cmp: % Complete | Completeness of the sequence (as a percentage). [NOTE: only sequences that are more than 90% complete and publicly available are currently included in this RDBMS.] | |||||||||||||||||||||||||||||||||||
| O: Open Reading Frame |
Indicates the presence or absence of an Open Reading Frame of at least 500 nucleotides in an intron sequence. More details about the designations are available.
|
|||||||||||||||||||||||||||||||||||
| AccNum: Accession Number | GenBank Accession Number [NCBI-National Center for Biotechnology Information]. Some sequences are assigned multiple accession numbers. By default, the RDBMS output displays only a single accession number for a sequence. If more accession numbers exist, m will appear next to the visible accession number. Click on m to see all of the accession numbers associated with that row. Click on s to bring you back to a single viewable accession number. | |||||||||||||||||||||||||||||||||||
| Only View Records w/Sec. Struct. Diagrams |
Search exclusively for sequence records with Secondary Structure diagrams.
NOTE: Our secondary structures are available in several formats: PostScript, PDF, and a simple text file containing the sequence and base pairing information. The structure diagrams can be obtained by either browsing the database or using the Data Retrieval Page to download many files at once. StrDiag: Secondary Structure DiagramsRDBMS Display of Secondary Structure Diagram Information:The RDBMS presents an abbreviated form of each secondary structure's full file name to conserve screen space. The following abbreviations are used: Diagram Version: the meaning of the letter is dependent upon the molecule, as follows:
Molecule:
Legend for Secondary Structure Diagrams:
More About Secondary Structure File Names:Filenames for the structure diagrams have the following syntax:
"Intron position (rRNA) OR number (other)" (shown in boldface above) is defined based upon the exon:
File name examples:
For a more detailed list, see "Codes used in Structure File Names." |
|||||||||||||||||||||||||||||||||||
| (.PS): PostScript | These secondary structure files are in PostScript format. PostScript files can be viewed with several different PostScript previewers (such as GhostScript). A GhostScript FAQ is available. A general description of PostScript is available in the document "A First Guide to PostScript". | |||||||||||||||||||||||||||||||||||
| (.PDF): Portable Document Format | Graphics files in this format can be viewed with the program Adobe Reader. | |||||||||||||||||||||||||||||||||||
| (.BPSEQ): BasePair/SEQuence Information |
Basepair and sequence information is presented in a text-based format. Each row presents information for one nucleotide in the sequence; the first field is the position number, the second field is the nucleotide at that position, and the third field contains the position number of its basepair partner (or 0 when unpaired). All secondary and tertiary basepairs are represented in this file, but base triples are not.
For 23S rRNA structures, a single BPSEQ file that contains all of the pairing information is available. Both the "235" and "233" links point to this single file. Secondary structure diagrams were interactively generated with the X-Windows-based graphics program "XRNA", developed by B. Weiser and H. Noller, University of California at Santa Cruz. |
|||||||||||||||||||||||||||||||||||
| (.ALDEN): ALDEN secondary structure format |
The Alden format (named for its developer, Matthew Alden) is a representation of RNA secondary structure. Each position in a sequence is grouped into a structural element segment, relates those segments into their parent structural elements, and shows the connectivity between structural elements. Each nucleotide in the sequence maps to one and only one structural element segment.
The current version of the Alden format presents each structural element segment, in 5' to 3' order, on one line containing 53 comma-delimited values (many of which are empty; this is a sparse format). See "Fields" (below) for details. The example provided below is from Escherichia coli 5S rRNA (Genbank Accession Number V00336). It contains examples of six of the eight types of structural element (lacks HELIX-KNOT and FREE). Structure Element Types (8):
Fields:
Example (click image for full-size version): 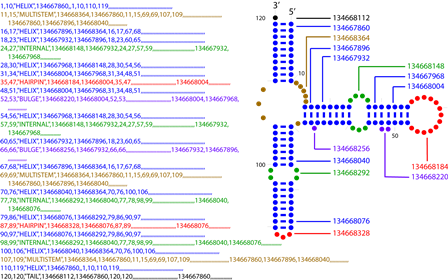
|
|||||||||||||||||||||||||||||||||||
| (.RNAML): RNA Markup Language |
RNAML is a markup language syntax designed for the exchange of RNA sequence and structure information. The official documentation for RNAML is located at the RNAML website.
The (.NOPBPSEQ) variant format excludes pseudoknot helices (to support programs which cannot accommodate pseudoknots). |
|||||||||||||||||||||||||||||||||||
| (.CT): Connect Table (.NOPCT): NO Pseudoknots CT |
The Connect Table format was designed to relate sequence and base pair information as input to structure-drawing programs. This text-based format presents the nucleotide number, nucleotide, 5' neighbor, 3' neighbor, pairing partner number (0 if unpaired), and original sequence nucleotide number (same as nucleotide number for all files including only one sequence). The official documentation for the CT format is located at the Mfold manual.
The (.NOPBPSEQ) variant format excludes pseudoknot helices (to support programs which cannot accommodate pseudoknots). |
|||||||||||||||||||||||||||||||||||
| (.BRACKET): Vienna Dot-BRACKET Notation | Dot-Bracket Notation represents pseudoknot-free sequences by a string of parens (open and close for the 5' and 3' nucleotides of a base-pair, respectively) and periods (unpaired nucleotides). A formal description of Dot-Bracket Notation is located at the Vienna RNA Webservers Help Page. | |||||||||||||||||||||||||||||||||||
| Results / Page | Number of sequence and/or secondary structure diagram records per page. The larger the number, the greater the time required to create and display the html page. | |||||||||||||||||||||||||||||||||||
| All Data Fields: | SEARCH ALL: Search for words or characters that occur in any attribute field. | |||||||||||||||||||||||||||||||||||
| Search Value (Examples) | Here is where the user specifies what they are searching for. Since this is a relational database, specifying certain attributes will limit the values for other attributes. A few examples of searches are listed here.
|
|||||||||||||||||||||||||||||||||||
| S: Sort Order | Sort the output for each attribute; a,b,c, ... -> z, 1,2,3, ... -> $. The user can specify the order that the attributes are sorted. The default sort order is for "Phylogenetic Classification" to be sorted first, followed by "Organism", "Cell Location", and then "RNA Class". For your convenience, the default sort order is displayed on the buttons in the Sort Order column. This order can be changed by clicking on the buttons in the Sort Order column. The first button clicked is sorted first, the second button clicked is sorted second, and so forth. The "Sort Reset" button restores the original default sort order. | |||||||||||||||||||||||||||||||||||
| R: Reverse Sort Order | The order of a sort for each sortable attribute can be inverted (e.g. z,y,x, ... -> a instead of a,b,c, ... -> z) by clicking on the appropriate button. | |||||||||||||||||||||||||||||||||||
| V: Values | Shows possible values for that attribute in the right frame window. A specific value is inserted into the search value box when that value in the right frame is clicked. | |||||||||||||||||||||||||||||||||||
| Comment: | May include other information about the sequence which does not fall into another category. | |||||||||||||||||||||||||||||||||||
| Color Coding: | Enabling this option causes the output of your search to display all entries for one organisms with the same background color.
Example: 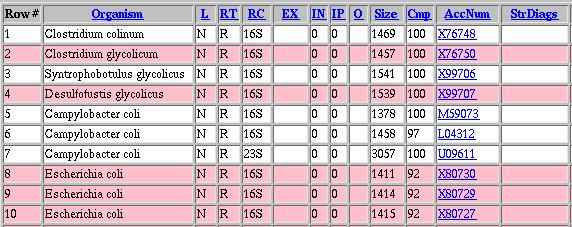
|
|||||||||||||||||||||||||||||||||||
